
Not wrapping but folding: Bacteria also organise their DNA (but they do it a bit differently)
Some bacteria, it turns out, have proteins much like ours that organise the DNA in their cells. They just do it a bit differently. This is revealed by new research from biochemists at the Leiden Institute of Chemistry and the Max Planck Institute for Biology. The discovery helps us better understand how bacteria organise their DNA and provides new insights into the evolution of these kinds of proteins.
‘That doesn’t quite add up,’ thought Professor Remus Dame when he read the publication from a group of fellow researchers. ‘They had found a protein structure similar to the one defined by my colleagues Birte Hernandez Alvarez anf Vikram Alva at the Max Planck Institute for Biology in Tübingen, Germany, who had also discovered the protein a few years ago. However, the way they claimed this protein binds to DNA seemed very illogical to me.’
Safe and neat DNA storage
Dame and his colleagues studied a special type of protein: histones. Histones play a crucial role in organising DNA in the cells of eukaryotes (cells with a nucleus) and archaea (single-celled organisms without a nucleus, see infobox). Dame: ‘DNA molecules are very long and contain crucial information for the cell. To store the DNA safely and compactly, a cell tightly winds the DNA strands around “beads” made of histones.’ By twisting the DNA around these histone beads in a specific way, the cell can also regulate which genes are accessible for transcription, thereby regulating their expression.
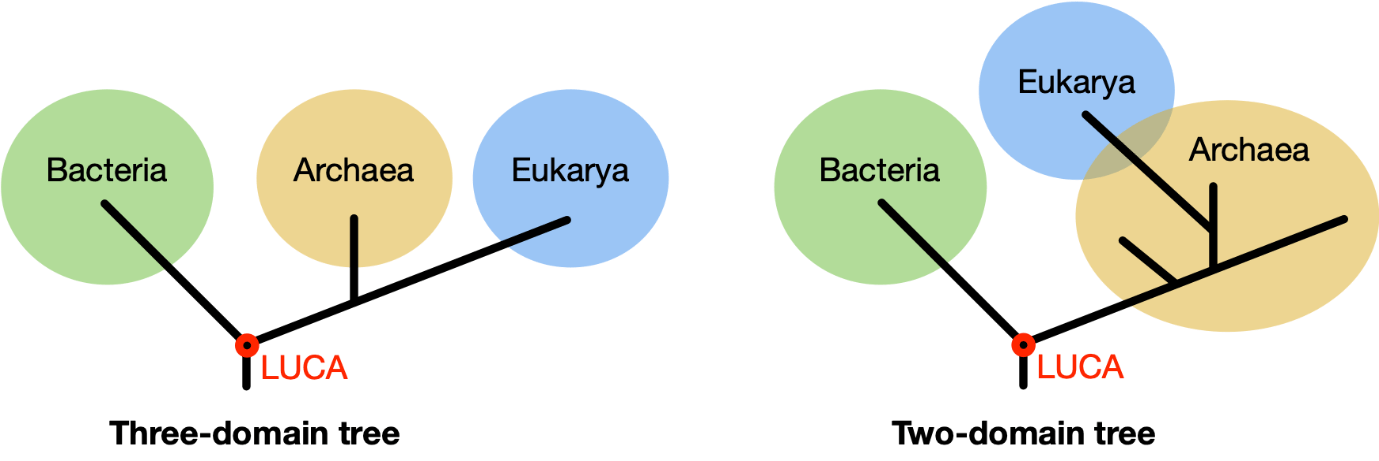
The domains of life
The most basic organisms on Earth are single-celled and have no nucleus. It wasn’t until around 1970 that it was discovered that these organisms form two different groups, namely bacteria and archaea. Superficially, they look similar, but their biochemistry is fundamentally different, which is why we consider them as two separate domains of life. Originally, organisms with cells that have a nucleus, the eukaryotes, including humans, were classified in a third domain. According to recent insights, however, there are only two domains of life, and eukaryotes belong to the archaea.
‘The exact opposite of what you'd expect’
For the first time, both the other research group and the colleagues from the Max Planck Institute described a histone protein in a bacterium. ‘That has not been done before,’ says Dame. ‘The DNA code and structure resembled a simplified version of “our” - human- histones. The question was: do they also have the same function?’ According to the competing group, they do not; they described a protein that wraps around the bacterial DNA and then stretches it out. Dame: ‘Exactly the opposite of what you would expect. That’s why I contacted our colleagues in Germany. Yimin Hu, a PhD candidate at the Max Planck Instiute, solved the structure of the protein bound to DNA, and we knew how to determine the protein’s function. And then everything came together beautifully.’
‘It actually makes the DNA compact’
Dame’s team conducted extensive biochemical analyses and single-molecule experiments. In these experiments, individual molecules are studied instead of large numbers at once. Dame: ‘This allowed us to show that this protein does exactly what you would expect: it binds to bacterial DNA in a very different way and makes it more compact.’

Not wrapping but folding
But this happens in a different way than in other life forms. In eukaryotes, histones form structures consisting of eight units. Together, they form a protein ball around which the DNA is wrapped. This also happens in archaea, but here the number of units is infinitely large, resulting in rod-shaped structures. In bacteria, it turns out to be once again quite different: the proteins form two units that do not wrap the DNA but bend it to make it compact.
Making the invisible visible
DNA is too small to see with the naked eye. So how do you study the effect of a protein on that DNA? The researchers came up with a simple but ingenious method. They add histone proteins to individual DNA strands and observe what happens. The setup is quite simple: one end of the DNA strand is fixed to a glass plate, while the other end hangs freely in the water, like an inverted pendulum. At this loose end, a plastic bead is attached, which you can see with the help of a simple microscope.

Because the beads are attached to the bottom, their range of movement is limited. Then you add a protein. PhD candidate Samuel Schwab explains: ‘The movement of the beads then tells us what happens to the DNA. If we add a protein that makes the DNA more compact, like our histone protein, we see that the beads start moving less. If the DNA became longer, you would see more movement. This way, you can indirectly gather information about the function of a protein.’
Emerging early in evolution
The research teaches us more about the functioning of histones in bacteria but also sheds new light on the early evolution of these crucial proteins. The fact that a simple form of histones already exists in some bacteria suggests that they emerged early in evolution. ‘It’s difficult to pinpoint exactly,’ says Dame. ‘But our discovery shows that bacterial histones may be an early, basic form of the more complex protein balls we find in eukaryotes and archaea. Even simple forms of life apparently already used sophisticated mechanisms to manage their genes. This way, bit by bit, we gain a deeper understanding of fundamental similarities and differences between organisms in our evolutionary tree.’
Scientific publication
Yimin Hu, Samuel Schwab, Silvia Deiss, Pedro Escudeiro, Thor van Heesch, Joe D Joiner, Jocelyne Vreede, Marcus D Hartmann, Andrei N Lupas, Birte Hernandez Alvarez, Vikram Alva, Remus T Dame, Bacterial histone HBb from Bdellovibrio bacteriovorus compacts DNA by bending, Nucleic Acids Research, 2024;, gkae485, https://doi.org/10.1093/nar/gkae485
