
A computational tool that will transform bacterial genome analysis
Whether a microbe is beneficial or harmful to a plant can now be predicted with high accuracy thanks to bacLIFE. This bioinformatic tool with an intuitive interface makes it much easier to unlock the secrets of bacterial genomes. A group of Leiden biologists presented it in Nature Communications.
‘Biologists often are afraid when they have to perform computational biology.’ Víctor Carrión emphasises the importance of bioinformatics in current biology. ‘PhD students sometimes spend their entire first year trying to get familiar with it.’ For example, most scientists need to be able to navigate various databases that show which genes are responsible for specific bacterial traits.
This tool welcomes researchers of all backgrounds
‘Anyone can freely screen any bacterial genome with just a few clicks’
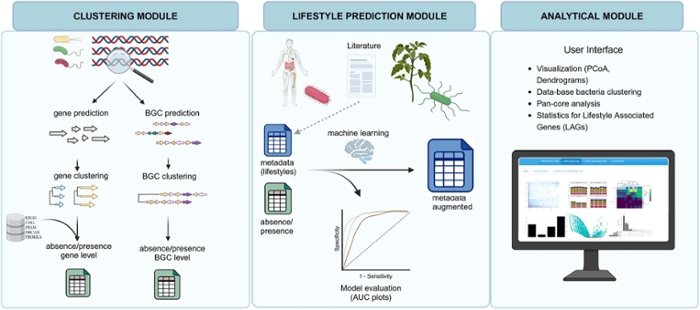
While existing bioinformatics pipelines often demand advanced computational expertise, the software tool bacLIFE takes a different approach. Designed to be accessible to the scientific community, it welcomes researchers of all backgrounds into the captivating world of bacterial genomics. ‘Anyone can freely screen any bacterial genome with just a few clicks. BacLIFE then provides a table after a few days of computation, predicting the proteins encoded in a particular genome, and which of those might be relevant for the bacterium’s pathogenic or beneficial lifestyle. With our tool, more and more genes are being identified from what we call the dark matter.’
The dark matter in the bacterial genome
Bacteria possess around three to four thousand genes. The function of about half of them is still unknown. ‘That is often referred to as the dark matter in the bacterial genomes,’ says Carrion. Some of those genes can potentially determine the bacterial lifestyle or their functions in a particular environment.

PhD candidate Guillermo Guerrero-Egido built the algorithm bacLIFE, using as input 16,856 genomes of bacteria from the genera (Para)burkholderia and Pseudomonas. BacLIFE predicted hundreds of genes that may encode proteins involved in bacterial traits that are pathogenic to plants.

They systematically switched off the genes
To test if the predictions of the algorithm were correct, his colleagues Kevin Bretscher and Adrian Pintado performed experiments. They selected fourteen genes that the algorithm predicted as involved in pathogenic lifestyle for rice plants. One by one, they systematically switched off these genes in bacteria by targeted mutagenesis. Using fourteen mutants, they infected rice plants and identified six genes that were involved in causing plant diseases. Thus, the utility of bacLIFE was proven.
The use of bacLIFE is now being tested in various of the Carrion lab projects. Bretscher conducted his time-consuming lab tests to verify bacLIFE’s predictions as a master’s student. Now, as a PhD candidate, he is investigating how bacteria can help plants to better survive in high salinity environments. With the help of bacLIFE. In other projects, the role of bacteria in the alleviation of plant drought stress is being studied. Carrión: ‘We are mainly focusing on agriculture, but bacLIFE could also be used for medical and biotechnological purposes.’
A new outbreak of bacterial disease
BacLIFE is currently tailored to the relationship between bacteria and plants, but if the programme also gains knowledge about, for example, bacteria-animal interactions, it would be useful in that area as well. Carrión: ‘If a new outbreak of a bacterial disease would emerge, researchers could sequence the pathogen’s genome and run it through bacLIFE. Then, they could quickly and precisely search for the genes and proteins that cause the disease, or for genes involved in resistance to antibiotics.’
Adjust the software and ‘feed’ it with genomes
Carríon sees even more possibilities: ‘With the help of bacLIFE, you can also more easily find out how to make bacteria produce a certain natural product, or perform a certain reaction in a bioreactor.’ Researchers can adjust the open-source software as needed and ‘feed’ it with genomes.
Using bacLIFE offers numerous possibilities that require relatively little time investment
Using bacLIFE offers numerous possibilities that require relatively little time investment for researchers. However, Guerrero-Egido and Bretscher put in a lot of hours for this result. Guerrero-Egido puzzled over the programme for a long time: ‘In the beginning, it gave far too many results, which were useless. We had to learn how to filter the results, But which criteria should we use for selection? I had to put a lot of effort into that.’
Quite a few drama meetings
Bretscher, who did the lab work with bacteria and plants, didn’t get his results easily either. ‘Not all bacteria are equally accessible to mutation techniques. It also took a lot of effort to get the cultivar of rice seeds used for these studies in the Netherlands. We had to make many changes to existing experimental protocols for it to work.’
The researchers had quite a few ‘drama meetings,’ they say. But when Bretscher saw how plants with a mutated bacterium grew much better than plants with the original, that made everything worthwhile. Carríon: ‘We’ve never seen him with such a big smile as then.’
Text: Rianne Lindhout
